 在 TensorFlow.org 上查看 在 TensorFlow.org 上查看
|
 在 Google Colab 中运行 在 Google Colab 中运行
|
 在 GitHub 上查看源代码 在 GitHub 上查看源代码
|
 下载笔记本 下载笔记本
|
潜在变量模型试图捕获高维数据中的隐藏结构。例如主成分分析 (PCA) 和因子分析。高斯过程是“非参数”模型,可以灵活地捕获局部相关结构和不确定性。高斯过程潜在变量模型 (Lawrence, 2004) 结合了这些概念。
背景:高斯过程
高斯过程是任何随机变量的集合,使得任何有限子集上的边际分布都是多元正态分布。有关回归环境下 GP 的详细介绍,请查看 TensorFlow Probability 中的高斯过程回归。
我们使用所谓的索引集来标记 GP 包含的集合中的每个随机变量。在有限索引集的情况下,我们只得到一个多元正态分布。但是,当我们考虑无限集合时,GP 最有趣。在像 \(\mathbb{R}^D\) 这样的索引集的情况下,我们为\(D\) 维空间中的每个点都有一个随机变量,GP 可以被认为是随机函数上的分布。从这样的 GP 中进行一次抽取,如果可以实现,将为 \(\mathbb{R}^D\) 中的每个点分配一个(联合正态分布的)值。在这个 colab 中,我们将重点关注一些 \(\mathbb{R}^D\) 上的 GP。
正态分布完全由其一阶和二阶统计量决定 - 事实上,定义正态分布的一种方法是,其高阶累积量都为零。对于 GP 也是如此:我们通过描述均值和协方差* 来完全指定一个 GP。回想一下,对于有限维多元正态分布,均值是一个向量,协方差是一个平方、对称正定矩阵。在无限维 GP 中,这些结构推广到定义在索引集每个点的均值函数 \(m : \mathbb{R}^D \to \mathbb{R}\),以及协方差“核”函数 \(k : \mathbb{R}^D \times \mathbb{R}^D \to \mathbb{R}\)。核函数需要是 正定 的,这本质上意味着,限制在有限点集上,它会产生一个正定矩阵。
GP 的大部分结构都源于其协方差核函数 - 此函数描述了采样函数的值在相邻(或不太相邻)点上的变化方式。不同的协方差函数会鼓励不同程度的平滑度。一个常用的核函数是“指数二次”(也称为“高斯”、“平方指数”或“径向基函数”),\(k(x, x') = \sigma^2 e^{(x - x^2) / \lambda^2}\)。其他示例在 David Duvenaud 的 核食谱页面 上以及规范文本 机器学习中的高斯过程 中概述。
* 对于无限索引集,我们还需要一个一致性条件。由于 GP 的定义是基于有限边缘分布的,我们必须要求这些边缘分布在无论以何种顺序取边缘分布时都是一致的。这是一个关于随机过程理论中比较高级的话题,超出了本教程的范围;总之,最终结果是好的!
应用 GP:回归和潜在变量模型
我们可以使用 GP 的一种方法是进行回归:给定一组以输入 \(\{x_i\}_{i=1}^N\)(索引集的元素)和观测值 \(\{y_i\}_{i=1}^N\) 形式的观测数据,我们可以使用这些数据在新的点集 \(\{x_j^*\}_{j=1}^M\) 上形成后验预测分布。由于所有分布都是高斯分布,因此这归结为一些简单的线性代数(但请注意:所需的计算在数据点数方面具有三次运行时间,并且需要在数据点数方面具有二次空间 - 这是使用 GP 的主要限制因素,目前许多研究都集中在计算上可行的精确后验推断替代方案上)。我们在 TFP colab 中的 GP 回归 中更详细地介绍了 GP 回归。
我们可以使用 GP 的另一种方法是作为潜在变量模型:给定一组高维观测值(例如,图像),我们可以假设一些低维潜在结构。我们假设,在给定潜在结构的情况下,大量输出(图像中的像素)彼此独立。此模型中的训练包括
- 优化模型参数(内核函数参数以及例如观测噪声方差),以及
- 为每个训练观测值(图像)找到索引集中相应的点位置。所有优化都可以通过最大化数据的边缘对数似然来完成。
导入
import numpy as np
import tensorflow as tf
import tf_keras
import tensorflow_probability as tfp
tfd = tfp.distributions
tfk = tfp.math.psd_kernels
%pylab inline
Populating the interactive namespace from numpy and matplotlib
加载 MNIST 数据
# Load the MNIST data set and isolate a subset of it.
(x_train, y_train), (_, _) = tf_keras.datasets.mnist.load_data()
N = 1000
small_x_train = x_train[:N, ...].astype(np.float64) / 256.
small_y_train = y_train[:N]
Downloading data from https://storage.googleapis.com/tensorflow/tf-keras-datasets/mnist.npz 11493376/11490434 [==============================] - 0s 0us/step 11501568/11490434 [==============================] - 0s 0us/step
准备可训练变量
我们将共同训练 3 个模型参数以及潜在输入。
# Create some trainable model parameters. We will constrain them to be strictly
# positive when constructing the kernel and the GP.
unconstrained_amplitude = tf.Variable(np.float64(1.), name='amplitude')
unconstrained_length_scale = tf.Variable(np.float64(1.), name='length_scale')
unconstrained_observation_noise = tf.Variable(np.float64(1.), name='observation_noise')
# We need to flatten the images and, somewhat unintuitively, transpose from
# shape [100, 784] to [784, 100]. This is because the 784 pixels will be
# treated as *independent* conditioned on the latent inputs, meaning we really
# have a batch of 784 GP's with 100 index_points.
observations_ = small_x_train.reshape(N, -1).transpose()
# Create a collection of N 2-dimensional index points that will represent our
# latent embeddings of the data. (Lawrence, 2004) prescribes initializing these
# with PCA, but a random initialization actually gives not-too-bad results, so
# we use this for simplicity. For a fun exercise, try doing the
# PCA-initialization yourself!
init_ = np.random.normal(size=(N, 2))
latent_index_points = tf.Variable(init_, name='latent_index_points')
构建模型和训练操作
# Create our kernel and GP distribution
EPS = np.finfo(np.float64).eps
def create_kernel():
amplitude = tf.math.softplus(EPS + unconstrained_amplitude)
length_scale = tf.math.softplus(EPS + unconstrained_length_scale)
kernel = tfk.ExponentiatedQuadratic(amplitude, length_scale)
return kernel
def loss_fn():
observation_noise_variance = tf.math.softplus(
EPS + unconstrained_observation_noise)
gp = tfd.GaussianProcess(
kernel=create_kernel(),
index_points=latent_index_points,
observation_noise_variance=observation_noise_variance)
log_probs = gp.log_prob(observations_, name='log_prob')
return -tf.reduce_mean(log_probs)
trainable_variables = [unconstrained_amplitude,
unconstrained_length_scale,
unconstrained_observation_noise,
latent_index_points]
optimizer = tf_keras.optimizers.Adam(learning_rate=1.0)
@tf.function(autograph=False, jit_compile=True)
def train_model():
with tf.GradientTape() as tape:
loss_value = loss_fn()
grads = tape.gradient(loss_value, trainable_variables)
optimizer.apply_gradients(zip(grads, trainable_variables))
return loss_value
训练并绘制生成的潜在嵌入
# Initialize variables and train!
num_iters = 100
log_interval = 20
lips = np.zeros((num_iters, N, 2), np.float64)
for i in range(num_iters):
loss = train_model()
lips[i] = latent_index_points.numpy()
if i % log_interval == 0 or i + 1 == num_iters:
print("Loss at step %d: %f" % (i, loss))
Loss at step 0: 1108.121688 Loss at step 20: -159.633761 Loss at step 40: -263.014394 Loss at step 60: -283.713056 Loss at step 80: -288.709413 Loss at step 99: -289.662253
绘制结果
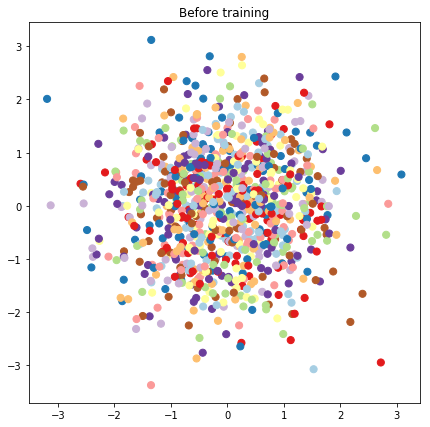
# Plot the latent locations before and after training
plt.figure(figsize=(7, 7))
plt.title("Before training")
plt.grid(False)
plt.scatter(x=init_[:, 0], y=init_[:, 1],
c=y_train[:N], cmap=plt.get_cmap('Paired'), s=50)
plt.show()
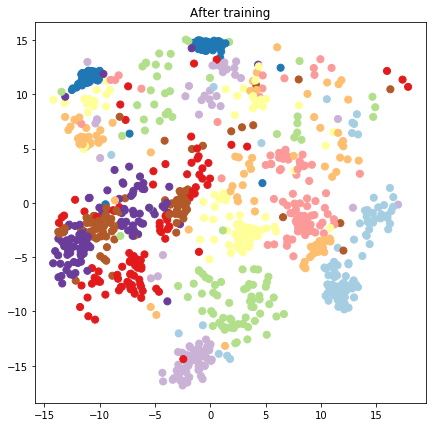
plt.figure(figsize=(7, 7))
plt.title("After training")
plt.grid(False)
plt.scatter(x=lips[-1, :, 0], y=lips[-1, :, 1],
c=y_train[:N], cmap=plt.get_cmap('Paired'), s=50)
plt.show()
构建预测模型和采样操作
# We'll draw samples at evenly spaced points on a 10x10 grid in the latent
# input space.
sample_grid_points = 10
grid_ = np.linspace(-4, 4, sample_grid_points).astype(np.float64)
# Create a 10x10 grid of 2-vectors, for a total shape [10, 10, 2]
grid_ = np.stack(np.meshgrid(grid_, grid_), axis=-1)
# This part's a bit subtle! What we defined above was a batch of 784 (=28x28)
# independent GP distributions over the input space. Each one corresponds to a
# single pixel of an MNIST image. Now what we'd like to do is draw 100 (=10x10)
# *independent* samples, each one separately conditioned on all the observations
# as well as the learned latent input locations above.
#
# The GP regression model below will define a batch of 784 independent
# posteriors. We'd like to get 100 independent samples each at a different
# latent index point. We could loop over the points in the grid, but that might
# be a bit slow. Instead, we can vectorize the computation by tacking on *even
# more* batch dimensions to our GaussianProcessRegressionModel distribution.
# In the below grid_ shape, we have concatentaed
# 1. batch shape: [sample_grid_points, sample_grid_points, 1]
# 2. number of examples: [1]
# 3. number of latent input dimensions: [2]
# The `1` in the batch shape will broadcast with 784. The final result will be
# samples of shape [10, 10, 784, 1]. The `1` comes from the "number of examples"
# and we can just `np.squeeze` it off.
grid_ = grid_.reshape(sample_grid_points, sample_grid_points, 1, 1, 2)
# Create the GPRegressionModel instance which represents the posterior
# predictive at the grid of new points.
gprm = tfd.GaussianProcessRegressionModel(
kernel=create_kernel(),
# Shape [10, 10, 1, 1, 2]
index_points=grid_,
# Shape [1000, 2]. 1000 2 dimensional vectors.
observation_index_points=latent_index_points,
# Shape [784, 1000]. A batch of 784 1000-dimensional observations.
observations=observations_)
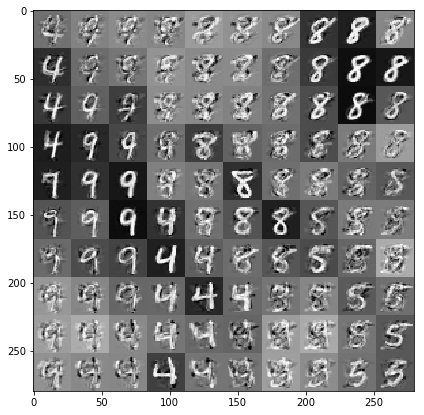
根据数据和潜在嵌入绘制样本
我们在潜在空间中二维网格上的 100 个点进行采样。
samples = gprm.sample()
# Plot the grid of samples at new points. We do a bit of tweaking of the samples
# first, squeezing off extra 1-shapes and normalizing the values.
samples_ = np.squeeze(samples.numpy())
samples_ = ((samples_ -
samples_.min(-1, keepdims=True)) /
(samples_.max(-1, keepdims=True) -
samples_.min(-1, keepdims=True)))
samples_ = samples_.reshape(sample_grid_points, sample_grid_points, 28, 28)
samples_ = samples_.transpose([0, 2, 1, 3])
samples_ = samples_.reshape(28 * sample_grid_points, 28 * sample_grid_points)
plt.figure(figsize=(7, 7))
ax = plt.subplot()
ax.grid(False)
ax.imshow(-samples_, interpolation='none', cmap='Greys')
plt.show()
结论
我们简要介绍了高斯过程潜在变量模型,并展示了如何在几行 TF 和 TF Probability 代码中实现它。
