 在 TensorFlow.org 上查看 在 TensorFlow.org 上查看
|
 在 Google Colab 中运行 在 Google Colab 中运行
|
 在 GitHub 上查看 在 GitHub 上查看
|
 下载笔记本 下载笔记本
|
在本 Colab 中,您将学习如何直接检查和创建模型的结构。我们假设您熟悉 初学者 和 中级 Colab 中介绍的概念。
在本 Colab 中,您将
训练一个随机森林模型并以编程方式访问其结构。
手动创建一个随机森林模型并将其用作经典模型。
设置
# Install TensorFlow Decision Forests.pip install tensorflow_decision_forests# Use wurlitzer to show the training logs.pip install wurlitzer
import os
# Keep using Keras 2
os.environ['TF_USE_LEGACY_KERAS'] = '1'
import tensorflow_decision_forests as tfdf
import numpy as np
import pandas as pd
import tensorflow as tf
import tf_keras
import matplotlib.pyplot as plt
import math
import collections
隐藏的代码单元格限制了 Colab 中的输出高度。
训练一个简单的随机森林
我们像在 初学者 Colab 中一样训练一个随机森林
# Download the dataset
!wget -q https://storage.googleapis.com/download.tensorflow.org/data/palmer_penguins/penguins.csv -O /tmp/penguins.csv
# Load a dataset into a Pandas Dataframe.
dataset_df = pd.read_csv("/tmp/penguins.csv")
# Show the first three examples.
print(dataset_df.head(3))
# Convert the pandas dataframe into a tf dataset.
dataset_tf = tfdf.keras.pd_dataframe_to_tf_dataset(dataset_df, label="species")
# Train the Random Forest
model = tfdf.keras.RandomForestModel(compute_oob_variable_importances=True)
model.fit(x=dataset_tf)
species island bill_length_mm bill_depth_mm flipper_length_mm \ 0 Adelie Torgersen 39.1 18.7 181.0 1 Adelie Torgersen 39.5 17.4 186.0 2 Adelie Torgersen 40.3 18.0 195.0 body_mass_g sex year 0 3750.0 male 2007 1 3800.0 female 2007 2 3250.0 female 2007 Warning: The `num_threads` constructor argument is not set and the number of CPU is os.cpu_count()=32 > 32. Setting num_threads to 32. Set num_threads manually to use more than 32 cpus. WARNING:absl:The `num_threads` constructor argument is not set and the number of CPU is os.cpu_count()=32 > 32. Setting num_threads to 32. Set num_threads manually to use more than 32 cpus. Use /tmpfs/tmp/tmpadwizz7x as temporary training directory Reading training dataset... Training dataset read in 0:00:03.574049. Found 344 examples. Training model... Model trained in 0:00:00.092571 Compiling model... [INFO 24-04-20 11:24:50.3886 UTC kernel.cc:1233] Loading model from path /tmpfs/tmp/tmpadwizz7x/model/ with prefix 59499fe5fa654879 [INFO 24-04-20 11:24:50.4047 UTC decision_forest.cc:734] Model loaded with 300 root(s), 5080 node(s), and 7 input feature(s). [INFO 24-04-20 11:24:50.4047 UTC abstract_model.cc:1344] Engine "RandomForestGeneric" built [INFO 24-04-20 11:24:50.4048 UTC kernel.cc:1061] Use fast generic engine Model compiled. <tf_keras.src.callbacks.History at 0x7fb16472dbe0>
请注意模型构造函数中的 compute_oob_variable_importances=True 超参数。此选项在训练期间计算袋外 (OOB) 变量重要性。这是随机森林模型的一种流行的 排列变量重要性。
计算 OOB 变量重要性不会影响最终模型,它会减慢大型数据集的训练速度。
检查模型摘要
%set_cell_height 300
model.summary()
<IPython.core.display.Javascript object>
Model: "random_forest_model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
=================================================================
Total params: 1 (1.00 Byte)
Trainable params: 0 (0.00 Byte)
Non-trainable params: 1 (1.00 Byte)
_________________________________________________________________
Type: "RANDOM_FOREST"
Task: CLASSIFICATION
Label: "__LABEL"
Input Features (7):
bill_depth_mm
bill_length_mm
body_mass_g
flipper_length_mm
island
sex
year
No weights
Variable Importance: INV_MEAN_MIN_DEPTH:
1. "flipper_length_mm" 0.440513 ################
2. "bill_length_mm" 0.438028 ###############
3. "bill_depth_mm" 0.299751 #####
4. "island" 0.295079 #####
5. "body_mass_g" 0.256534 ##
6. "sex" 0.225708
7. "year" 0.224020
Variable Importance: MEAN_DECREASE_IN_ACCURACY:
1. "bill_length_mm" 0.151163 ################
2. "island" 0.008721 #
3. "bill_depth_mm" 0.000000
4. "body_mass_g" 0.000000
5. "sex" 0.000000
6. "year" 0.000000
7. "flipper_length_mm" -0.002907
Variable Importance: MEAN_DECREASE_IN_AP_1_VS_OTHERS:
1. "bill_length_mm" 0.083305 ################
2. "island" 0.007664 #
3. "flipper_length_mm" 0.003400
4. "bill_depth_mm" 0.002741
5. "body_mass_g" 0.000722
6. "sex" 0.000644
7. "year" 0.000000
Variable Importance: MEAN_DECREASE_IN_AP_2_VS_OTHERS:
1. "bill_length_mm" 0.508510 ################
2. "island" 0.023487
3. "bill_depth_mm" 0.007744
4. "flipper_length_mm" 0.006008
5. "body_mass_g" 0.003017
6. "sex" 0.001537
7. "year" -0.000245
Variable Importance: MEAN_DECREASE_IN_AP_3_VS_OTHERS:
1. "island" 0.002192 ################
2. "bill_length_mm" 0.001572 ############
3. "bill_depth_mm" 0.000497 #######
4. "sex" 0.000000 ####
5. "year" 0.000000 ####
6. "body_mass_g" -0.000053 ####
7. "flipper_length_mm" -0.000890
Variable Importance: MEAN_DECREASE_IN_AUC_1_VS_OTHERS:
1. "bill_length_mm" 0.071306 ################
2. "island" 0.007299 #
3. "flipper_length_mm" 0.004506 #
4. "bill_depth_mm" 0.002124
5. "body_mass_g" 0.000548
6. "sex" 0.000480
7. "year" 0.000000
Variable Importance: MEAN_DECREASE_IN_AUC_2_VS_OTHERS:
1. "bill_length_mm" 0.108642 ################
2. "island" 0.014493 ##
3. "bill_depth_mm" 0.007406 #
4. "flipper_length_mm" 0.005195
5. "body_mass_g" 0.001012
6. "sex" 0.000480
7. "year" -0.000053
Variable Importance: MEAN_DECREASE_IN_AUC_3_VS_OTHERS:
1. "island" 0.002126 ################
2. "bill_length_mm" 0.001393 ###########
3. "bill_depth_mm" 0.000293 #####
4. "sex" 0.000000 ###
5. "year" 0.000000 ###
6. "body_mass_g" -0.000037 ###
7. "flipper_length_mm" -0.000550
Variable Importance: MEAN_DECREASE_IN_PRAUC_1_VS_OTHERS:
1. "bill_length_mm" 0.083122 ################
2. "island" 0.010887 ##
3. "flipper_length_mm" 0.003425
4. "bill_depth_mm" 0.002731
5. "body_mass_g" 0.000719
6. "sex" 0.000641
7. "year" 0.000000
Variable Importance: MEAN_DECREASE_IN_PRAUC_2_VS_OTHERS:
1. "bill_length_mm" 0.497611 ################
2. "island" 0.024045
3. "bill_depth_mm" 0.007734
4. "flipper_length_mm" 0.006017
5. "body_mass_g" 0.003000
6. "sex" 0.001528
7. "year" -0.000243
Variable Importance: MEAN_DECREASE_IN_PRAUC_3_VS_OTHERS:
1. "island" 0.002187 ################
2. "bill_length_mm" 0.001568 ############
3. "bill_depth_mm" 0.000495 #######
4. "sex" 0.000000 ####
5. "year" 0.000000 ####
6. "body_mass_g" -0.000053 ####
7. "flipper_length_mm" -0.000886
Variable Importance: NUM_AS_ROOT:
1. "flipper_length_mm" 157.000000 ################
2. "bill_length_mm" 76.000000 #######
3. "bill_depth_mm" 52.000000 #####
4. "island" 12.000000
5. "body_mass_g" 3.000000
Variable Importance: NUM_NODES:
1. "bill_length_mm" 778.000000 ################
2. "bill_depth_mm" 463.000000 #########
3. "flipper_length_mm" 414.000000 ########
4. "island" 342.000000 ######
5. "body_mass_g" 338.000000 ######
6. "sex" 36.000000
7. "year" 19.000000
Variable Importance: SUM_SCORE:
1. "bill_length_mm" 36515.793787 ################
2. "flipper_length_mm" 35120.434174 ###############
3. "island" 14669.408395 ######
4. "bill_depth_mm" 14515.446617 ######
5. "body_mass_g" 3485.330881 #
6. "sex" 354.201073
7. "year" 49.737758
Winner takes all: true
Out-of-bag evaluation: accuracy:0.976744 logloss:0.068949
Number of trees: 300
Total number of nodes: 5080
Number of nodes by tree:
Count: 300 Average: 16.9333 StdDev: 3.10197
Min: 11 Max: 31 Ignored: 0
----------------------------------------------
[ 11, 12) 6 2.00% 2.00% #
[ 12, 13) 0 0.00% 2.00%
[ 13, 14) 46 15.33% 17.33% #####
[ 14, 15) 0 0.00% 17.33%
[ 15, 16) 70 23.33% 40.67% ########
[ 16, 17) 0 0.00% 40.67%
[ 17, 18) 84 28.00% 68.67% ##########
[ 18, 19) 0 0.00% 68.67%
[ 19, 20) 46 15.33% 84.00% #####
[ 20, 21) 0 0.00% 84.00%
[ 21, 22) 30 10.00% 94.00% ####
[ 22, 23) 0 0.00% 94.00%
[ 23, 24) 13 4.33% 98.33% ##
[ 24, 25) 0 0.00% 98.33%
[ 25, 26) 2 0.67% 99.00%
[ 26, 27) 0 0.00% 99.00%
[ 27, 28) 2 0.67% 99.67%
[ 28, 29) 0 0.00% 99.67%
[ 29, 30) 0 0.00% 99.67%
[ 30, 31] 1 0.33% 100.00%
Depth by leafs:
Count: 2690 Average: 3.53271 StdDev: 1.06789
Min: 2 Max: 7 Ignored: 0
----------------------------------------------
[ 2, 3) 545 20.26% 20.26% ######
[ 3, 4) 747 27.77% 48.03% ########
[ 4, 5) 888 33.01% 81.04% ##########
[ 5, 6) 444 16.51% 97.55% #####
[ 6, 7) 62 2.30% 99.85% #
[ 7, 7] 4 0.15% 100.00%
Number of training obs by leaf:
Count: 2690 Average: 38.3643 StdDev: 44.8651
Min: 5 Max: 155 Ignored: 0
----------------------------------------------
[ 5, 12) 1474 54.80% 54.80% ##########
[ 12, 20) 124 4.61% 59.41% #
[ 20, 27) 48 1.78% 61.19%
[ 27, 35) 74 2.75% 63.94% #
[ 35, 42) 58 2.16% 66.10%
[ 42, 50) 85 3.16% 69.26% #
[ 50, 57) 96 3.57% 72.83% #
[ 57, 65) 87 3.23% 76.06% #
[ 65, 72) 49 1.82% 77.88%
[ 72, 80) 23 0.86% 78.74%
[ 80, 88) 30 1.12% 79.85%
[ 88, 95) 23 0.86% 80.71%
[ 95, 103) 42 1.56% 82.27%
[ 103, 110) 62 2.30% 84.57%
[ 110, 118) 115 4.28% 88.85% #
[ 118, 125) 115 4.28% 93.12% #
[ 125, 133) 98 3.64% 96.77% #
[ 133, 140) 49 1.82% 98.59%
[ 140, 148) 31 1.15% 99.74%
[ 148, 155] 7 0.26% 100.00%
Attribute in nodes:
778 : bill_length_mm [NUMERICAL]
463 : bill_depth_mm [NUMERICAL]
414 : flipper_length_mm [NUMERICAL]
342 : island [CATEGORICAL]
338 : body_mass_g [NUMERICAL]
36 : sex [CATEGORICAL]
19 : year [NUMERICAL]
Attribute in nodes with depth <= 0:
157 : flipper_length_mm [NUMERICAL]
76 : bill_length_mm [NUMERICAL]
52 : bill_depth_mm [NUMERICAL]
12 : island [CATEGORICAL]
3 : body_mass_g [NUMERICAL]
Attribute in nodes with depth <= 1:
250 : bill_length_mm [NUMERICAL]
244 : flipper_length_mm [NUMERICAL]
183 : bill_depth_mm [NUMERICAL]
170 : island [CATEGORICAL]
53 : body_mass_g [NUMERICAL]
Attribute in nodes with depth <= 2:
462 : bill_length_mm [NUMERICAL]
320 : flipper_length_mm [NUMERICAL]
310 : bill_depth_mm [NUMERICAL]
287 : island [CATEGORICAL]
162 : body_mass_g [NUMERICAL]
9 : sex [CATEGORICAL]
5 : year [NUMERICAL]
Attribute in nodes with depth <= 3:
669 : bill_length_mm [NUMERICAL]
410 : bill_depth_mm [NUMERICAL]
383 : flipper_length_mm [NUMERICAL]
328 : island [CATEGORICAL]
286 : body_mass_g [NUMERICAL]
32 : sex [CATEGORICAL]
10 : year [NUMERICAL]
Attribute in nodes with depth <= 5:
778 : bill_length_mm [NUMERICAL]
462 : bill_depth_mm [NUMERICAL]
413 : flipper_length_mm [NUMERICAL]
342 : island [CATEGORICAL]
338 : body_mass_g [NUMERICAL]
36 : sex [CATEGORICAL]
19 : year [NUMERICAL]
Condition type in nodes:
2012 : HigherCondition
378 : ContainsBitmapCondition
Condition type in nodes with depth <= 0:
288 : HigherCondition
12 : ContainsBitmapCondition
Condition type in nodes with depth <= 1:
730 : HigherCondition
170 : ContainsBitmapCondition
Condition type in nodes with depth <= 2:
1259 : HigherCondition
296 : ContainsBitmapCondition
Condition type in nodes with depth <= 3:
1758 : HigherCondition
360 : ContainsBitmapCondition
Condition type in nodes with depth <= 5:
2010 : HigherCondition
378 : ContainsBitmapCondition
Node format: NOT_SET
Training OOB:
trees: 1, Out-of-bag evaluation: accuracy:0.964286 logloss:1.28727
trees: 13, Out-of-bag evaluation: accuracy:0.94863 logloss:1.38235
trees: 29, Out-of-bag evaluation: accuracy:0.963526 logloss:0.698239
trees: 39, Out-of-bag evaluation: accuracy:0.958824 logloss:0.37345
trees: 54, Out-of-bag evaluation: accuracy:0.973837 logloss:0.171543
trees: 72, Out-of-bag evaluation: accuracy:0.97093 logloss:0.171775
trees: 82, Out-of-bag evaluation: accuracy:0.973837 logloss:0.168111
trees: 92, Out-of-bag evaluation: accuracy:0.976744 logloss:0.167506
trees: 113, Out-of-bag evaluation: accuracy:0.976744 logloss:0.170507
trees: 124, Out-of-bag evaluation: accuracy:0.976744 logloss:0.07406
trees: 135, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0739305
trees: 145, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0741686
trees: 155, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0738562
trees: 166, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0727146
trees: 177, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0721128
trees: 195, Out-of-bag evaluation: accuracy:0.976744 logloss:0.070882
trees: 205, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0705714
trees: 216, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0697382
trees: 231, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0695581
trees: 244, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0683962
trees: 255, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0693447
trees: 267, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0689024
trees: 279, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0694214
trees: 296, Out-of-bag evaluation: accuracy:0.976744 logloss:0.0691636
trees: 300, Out-of-bag evaluation: accuracy:0.976744 logloss:0.068949
请注意名称为 MEAN_DECREASE_IN_* 的多个变量重要性。
绘制模型
接下来,绘制模型。
随机森林是一个大型模型(此模型有 300 棵树和约 5k 个节点;请参阅上面的摘要)。因此,只绘制第一棵树,并将节点限制为深度 3。
tfdf.model_plotter.plot_model_in_colab(model, tree_idx=0, max_depth=3)
检查模型结构
模型结构和元数据可通过 make_inspector() 创建的 **检查器** 获取。
inspector = model.make_inspector()
对于我们的模型,可用的检查器字段是
[field for field in dir(inspector) if not field.startswith("_")]
['MODEL_NAME', 'dataspec', 'directory', 'evaluation', 'export_to_tensorboard', 'extract_all_trees', 'extract_tree', 'features', 'file_prefix', 'header', 'iterate_on_nodes', 'label', 'label_classes', 'metadata', 'model_type', 'num_trees', 'objective', 'specialized_header', 'task', 'training_logs', 'tuning_logs', 'variable_importances', 'winner_take_all_inference']
请记住查看 API 参考 或使用 ? 获取内置文档。
?inspector.model_type
一些模型元数据
print("Model type:", inspector.model_type())
print("Number of trees:", inspector.num_trees())
print("Objective:", inspector.objective())
print("Input features:", inspector.features())
Model type: RANDOM_FOREST Number of trees: 300 Objective: Classification(label=__LABEL, class=None, num_classes=3) Input features: ["bill_depth_mm" (1; #1), "bill_length_mm" (1; #2), "body_mass_g" (1; #3), "flipper_length_mm" (1; #4), "island" (4; #5), "sex" (4; #6), "year" (1; #7)]
evaluate() 是在训练期间计算的模型评估。用于此评估的数据集取决于算法。例如,它可以是验证数据集或袋外数据集。
inspector.evaluation()
Evaluation(num_examples=344, accuracy=0.9767441860465116, loss=0.06894904488784283, rmse=None, ndcg=None, aucs=None, auuc=None, qini=None)
变量重要性是
print(f"Available variable importances:")
for importance in inspector.variable_importances().keys():
print("\t", importance)
Available variable importances:
MEAN_DECREASE_IN_PRAUC_3_VS_OTHERS
MEAN_DECREASE_IN_PRAUC_1_VS_OTHERS
INV_MEAN_MIN_DEPTH
MEAN_DECREASE_IN_AUC_1_VS_OTHERS
MEAN_DECREASE_IN_AP_2_VS_OTHERS
MEAN_DECREASE_IN_AUC_3_VS_OTHERS
MEAN_DECREASE_IN_AUC_2_VS_OTHERS
MEAN_DECREASE_IN_AP_1_VS_OTHERS
NUM_AS_ROOT
NUM_NODES
MEAN_DECREASE_IN_PRAUC_2_VS_OTHERS
MEAN_DECREASE_IN_ACCURACY
SUM_SCORE
MEAN_DECREASE_IN_AP_3_VS_OTHERS
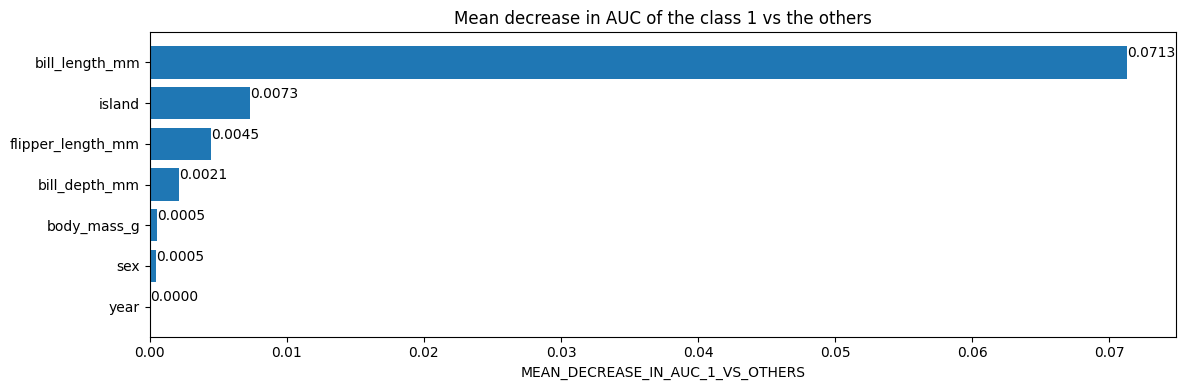
不同的变量重要性具有不同的语义。例如,一个特征的 **平均 AUC 降低** 为 0.05 意味着从训练数据集中删除此特征会降低/损害 AUC 5%。
# Mean decrease in AUC of the class 1 vs the others.
inspector.variable_importances()["MEAN_DECREASE_IN_AUC_1_VS_OTHERS"]
[("bill_length_mm" (1; #2), 0.0713061951754389),
("island" (4; #5), 0.007298519736842035),
("flipper_length_mm" (1; #4), 0.004505893640351366),
("bill_depth_mm" (1; #1), 0.0021244517543865804),
("body_mass_g" (1; #3), 0.0005482456140351033),
("sex" (4; #6), 0.00047971491228060437),
("year" (1; #7), 0.0)]
使用 Matplotlib 绘制检查器中的变量重要性
import matplotlib.pyplot as plt
plt.figure(figsize=(12, 4))
# Mean decrease in AUC of the class 1 vs the others.
variable_importance_metric = "MEAN_DECREASE_IN_AUC_1_VS_OTHERS"
variable_importances = inspector.variable_importances()[variable_importance_metric]
# Extract the feature name and importance values.
#
# `variable_importances` is a list of <feature, importance> tuples.
feature_names = [vi[0].name for vi in variable_importances]
feature_importances = [vi[1] for vi in variable_importances]
# The feature are ordered in decreasing importance value.
feature_ranks = range(len(feature_names))
bar = plt.barh(feature_ranks, feature_importances, label=[str(x) for x in feature_ranks])
plt.yticks(feature_ranks, feature_names)
plt.gca().invert_yaxis()
# TODO: Replace with "plt.bar_label()" when available.
# Label each bar with values
for importance, patch in zip(feature_importances, bar.patches):
plt.text(patch.get_x() + patch.get_width(), patch.get_y(), f"{importance:.4f}", va="top")
plt.xlabel(variable_importance_metric)
plt.title("Mean decrease in AUC of the class 1 vs the others")
plt.tight_layout()
plt.show()
最后,访问实际的树结构
inspector.extract_tree(tree_idx=0)
Tree(root=NonLeafNode(condition=(bill_length_mm >= 43.25; miss=True, score=0.5482327342033386), pos_child=NonLeafNode(condition=(island in ['Biscoe']; miss=True, score=0.6515106558799744), pos_child=NonLeafNode(condition=(bill_depth_mm >= 17.225584030151367; miss=False, score=0.027205035090446472), pos_child=LeafNode(value=ProbabilityValue([0.16666666666666666, 0.0, 0.8333333333333334],n=6.0), idx=7), neg_child=LeafNode(value=ProbabilityValue([0.0, 0.0, 1.0],n=104.0), idx=6), value=ProbabilityValue([0.00909090909090909, 0.0, 0.990909090909091],n=110.0)), neg_child=LeafNode(value=ProbabilityValue([0.0, 1.0, 0.0],n=61.0), idx=5), value=ProbabilityValue([0.005847953216374269, 0.3567251461988304, 0.6374269005847953],n=171.0)), neg_child=NonLeafNode(condition=(bill_depth_mm >= 15.100000381469727; miss=True, score=0.150658518075943), pos_child=NonLeafNode(condition=(flipper_length_mm >= 187.5; miss=True, score=0.036139510571956635), pos_child=LeafNode(value=ProbabilityValue([1.0, 0.0, 0.0],n=104.0), idx=4), neg_child=NonLeafNode(condition=(bill_length_mm >= 42.30000305175781; miss=True, score=0.23430533707141876), pos_child=LeafNode(value=ProbabilityValue([0.0, 1.0, 0.0],n=5.0), idx=3), neg_child=NonLeafNode(condition=(bill_length_mm >= 40.55000305175781; miss=True, score=0.043961383402347565), pos_child=LeafNode(value=ProbabilityValue([0.8, 0.2, 0.0],n=5.0), idx=2), neg_child=LeafNode(value=ProbabilityValue([1.0, 0.0, 0.0],n=53.0), idx=1), value=ProbabilityValue([0.9827586206896551, 0.017241379310344827, 0.0],n=58.0)), value=ProbabilityValue([0.9047619047619048, 0.09523809523809523, 0.0],n=63.0)), value=ProbabilityValue([0.9640718562874252, 0.03592814371257485, 0.0],n=167.0)), neg_child=LeafNode(value=ProbabilityValue([0.0, 0.0, 1.0],n=6.0), idx=0), value=ProbabilityValue([0.930635838150289, 0.03468208092485549, 0.03468208092485549],n=173.0)), value=ProbabilityValue([0.47093023255813954, 0.19476744186046513, 0.33430232558139533],n=344.0)), label_classes=None)
提取树效率不高。如果速度很重要,可以使用 iterate_on_nodes() 方法进行模型检查。此方法是模型所有节点的深度优先前序遍历迭代器。
以下示例计算每个特征的使用次数(这是一种结构变量重要性)。
# number_of_use[F] will be the number of node using feature F in its condition.
number_of_use = collections.defaultdict(lambda: 0)
# Iterate over all the nodes in a Depth First Pre-order traversals.
for node_iter in inspector.iterate_on_nodes():
if not isinstance(node_iter.node, tfdf.py_tree.node.NonLeafNode):
# Skip the leaf nodes
continue
# Iterate over all the features used in the condition.
# By default, models are "oblique" i.e. each node tests a single feature.
for feature in node_iter.node.condition.features():
number_of_use[feature] += 1
print("Number of condition nodes per features:")
for feature, count in number_of_use.items():
print("\t", feature.name, ":", count)
Number of condition nodes per features:
bill_length_mm : 778
bill_depth_mm : 463
flipper_length_mm : 414
island : 342
body_mass_g : 338
year : 19
sex : 36
手动创建模型
在本节中,您将手动创建一个小型随机森林模型。为了使其更易于操作,该模型将只包含一棵简单的树
3 label classes: Red, blue and green.
2 features: f1 (numerical) and f2 (string categorical)
f1>=1.5
├─(pos)─ f2 in ["cat","dog"]
│ ├─(pos)─ value: [0.8, 0.1, 0.1]
│ └─(neg)─ value: [0.1, 0.8, 0.1]
└─(neg)─ value: [0.1, 0.1, 0.8]
# Create the model builder
builder = tfdf.builder.RandomForestBuilder(
path="/tmp/manual_model",
objective=tfdf.py_tree.objective.ClassificationObjective(
label="color", classes=["red", "blue", "green"]))
每棵树都逐个添加。
# So alias
Tree = tfdf.py_tree.tree.Tree
SimpleColumnSpec = tfdf.py_tree.dataspec.SimpleColumnSpec
ColumnType = tfdf.py_tree.dataspec.ColumnType
# Nodes
NonLeafNode = tfdf.py_tree.node.NonLeafNode
LeafNode = tfdf.py_tree.node.LeafNode
# Conditions
NumericalHigherThanCondition = tfdf.py_tree.condition.NumericalHigherThanCondition
CategoricalIsInCondition = tfdf.py_tree.condition.CategoricalIsInCondition
# Leaf values
ProbabilityValue = tfdf.py_tree.value.ProbabilityValue
builder.add_tree(
Tree(
NonLeafNode(
condition=NumericalHigherThanCondition(
feature=SimpleColumnSpec(name="f1", type=ColumnType.NUMERICAL),
threshold=1.5,
missing_evaluation=False),
pos_child=NonLeafNode(
condition=CategoricalIsInCondition(
feature=SimpleColumnSpec(name="f2",type=ColumnType.CATEGORICAL),
mask=["cat", "dog"],
missing_evaluation=False),
pos_child=LeafNode(value=ProbabilityValue(probability=[0.8, 0.1, 0.1], num_examples=10)),
neg_child=LeafNode(value=ProbabilityValue(probability=[0.1, 0.8, 0.1], num_examples=20))),
neg_child=LeafNode(value=ProbabilityValue(probability=[0.1, 0.1, 0.8], num_examples=30)))))
完成树的写入
builder.close()
[INFO 24-04-20 11:24:54.9480 UTC kernel.cc:1233] Loading model from path /tmp/manual_model/tmp/ with prefix f938aac6d7ed44f5 [INFO 24-04-20 11:24:54.9483 UTC decision_forest.cc:734] Model loaded with 1 root(s), 5 node(s), and 2 input feature(s). [INFO 24-04-20 11:24:54.9483 UTC kernel.cc:1061] Use fast generic engine INFO:tensorflow:Assets written to: /tmp/manual_model/assets INFO:tensorflow:Assets written to: /tmp/manual_model/assets
现在,您可以像常规 Keras 模型一样打开模型,并进行预测
manual_model = tf_keras.models.load_model("/tmp/manual_model")
[INFO 24-04-20 11:24:56.1029 UTC kernel.cc:1233] Loading model from path /tmp/manual_model/assets/ with prefix f938aac6d7ed44f5 [INFO 24-04-20 11:24:56.1032 UTC decision_forest.cc:734] Model loaded with 1 root(s), 5 node(s), and 2 input feature(s). [INFO 24-04-20 11:24:56.1032 UTC kernel.cc:1061] Use fast generic engine
examples = tf.data.Dataset.from_tensor_slices({
"f1": [1.0, 2.0, 3.0],
"f2": ["cat", "cat", "bird"]
}).batch(2)
predictions = manual_model.predict(examples)
print("predictions:\n",predictions)
2/2 [==============================] - 1s 3ms/step predictions: [[0.1 0.1 0.8] [0.8 0.1 0.1] [0.1 0.8 0.1]]
访问结构
yggdrasil_model_path = manual_model.yggdrasil_model_path_tensor().numpy().decode("utf-8")
print("yggdrasil_model_path:",yggdrasil_model_path)
inspector = tfdf.inspector.make_inspector(yggdrasil_model_path)
print("Input features:", inspector.features())
yggdrasil_model_path: /tmp/manual_model/assets/ Input features: ["f1" (1; #1), "f2" (4; #2)]
当然,您可以绘制此手动构建的模型
tfdf.model_plotter.plot_model_in_colab(manual_model)
